|
- 品牌:Absin
- 产地:上海
- 货号:abs60541
- 价格: ¥2800/份
- 发布日期: 2023-09-14
- 更新日期: 2026-02-14
| 产地 | 上海 |
| 品牌 | Absin |
| 货号 | abs60541 |
| 用途 | - |
| 包装规格 | 100T |
| CAS编号 | |
| 纯度 | 内毒素水平:<0.1EU/ug% |
| 是否进口 | 否 |
产品描述:
The Baculovirus Quantitative Titration Kit serves as an extraction and titration system for fresh (less than 3 months old) budded baculovirus stocks based on AcMNPV baculoviral vectors. Ultra-sensitive TaqMan qPCR technique and accurate titrated virus stocks are employed in this kit, providing an alternatively time-saving method of titration up to 2-3 hours. Therefore, reliable multiplicity of infection (MOI) can be determined real-timely, substituting traditional plaque assays or antibody-based assays which usually take 3-4 days.
The primers and probe included in this kit are specific to gp64 that encodes the essential envelope protein GP64, and the sequences, concentration and ration are also optimized. Virus Lysis Buffer from this kit can degrade viral protein, releasing viral DNA that can be used directly as template for qPCR without further purification. Virus Standard, Positive Control (PC) and negative template control (NTC) should be set up every time of quantitative titration.
Components:
| Components | Amount | Storage |
| Virus Standard | AcMNPV Virus, 2×108 pfu/mL | -20℃ |
| irus Lysis Buffer | 1mL | -20℃ |
| Proteinase K* | 10×, 100uL | -20℃ |
| Primer & Probe Mix | Forward Primer & Reverse Primer & Probe, 240uL |
-20℃
IN THE DARK
|
| qPCR Low ROX mix? | 1.2mL | -20℃ |
| DNA Positive Control | 20ng/uL, 25uL | -20℃ |
| RNase-free Water | 1.5mL×3 | -20℃ |
* Add Proteinase K to Virus Lysis Buffer before use according to the number of your samples.
? Compatible to ABI Prism 7500/7500 Fast, QuantStudio 3/5/6 Flex/7 Flex System, ViiATM 7 System, Stratagene Mx3000/Mx3005P, Corbett Rotor Gene 3000, et. al.
使用方法:
Additional Materials Required:
Quantitative real-time PCR Thermocycler; Optical 96-well PCR plates or 8-well PCR strips with strip caps; Centrifuge for tubes, PCR strips or PCR plates
Baculoviral Titration Protocol
1. Centrifuge the medium from baculovirus-infected cells at 500g for 5mins. Transfer supernatant to a new 1.5 mL tube, discard cells and debris.
2. Dilute Virus Standard serially using the RNase-free water to generate standard serial log dilutions containing 6 points, 2×108pfu/mL, 2×107pfu/mL, 2×106pfu/mL, 2×105pfu/mL, 2×104pfu/mL, 2×103pfu/mL.
3. Dilute your unknown samples, if you suspect the titer to be great than 108pfu/mL.
4. Take 80uL standard serial dilutions and your unknown diluted (e.g., 1 in 5 or 1 in 10) or undiluted samples to PCR tubes, respectively. Centrifuge at 13000rpm for 5min, and then discard supernatant.
5. Re-suspend the virus pellet in 20uL Virus Lysis Buffer (add final 1x Proteinase K before use)in tips.
6. Run the following lysis program to extract virus DNA. The lysis can be used as templates for qPCR.
| Temperature | Time | |
| Step 1 | 55°C | 30min |
| Step 2 | 95°C | 5min |
| Step 3 | 55°C | 4min |
| Step 4 | 95°C | 1min |
| Step 5 | 55°C | 1min |
| Step 6 | 95°C | 30s |
| Step 7 | 20°C | Hold |
| Component | 20uL |
| qPCR mix(2×) | 10uL |
| Primer&Probe Mix | 2uL |
| Template | 2uL |
| RNase-free water | To 20uL |
| 2×108pfu/mL | 2×108pfu/mL | 2×108pfu/mL | Sample1 | Sample1 | Sample1 |
| 2×107pfu/mL | 2×107pfu/mL | 2×107pfu/mL | Sample2 | Sample2 | Sample2 |
| 2×106pfu/mL | 2×106pfu/mL | 2×106pfu/mL | Sample3 | Sample3 | Sample3 |
| 2×105pfu/mL | 2×105pfu/mL | 2×105pfu/mL | Sample4 | Sample4 | Sample4 |
| 2×104pfu/mL | 2×104pfu/mL | 2×104pfu/mL | Sample5 | Sample5 | Sample5 |
| 2×103pfu/mL | 2×103pfu/mL | 2×103pfu/mL | Sample6 | Sample6 | Sample6 |
| PC | PC | PC | ... | ... | ... |
| NTC | NTC | NTC | ... | ... | ... |
| Temperature | Time | Cycle |
| 95°C | 2min | 1× |
| 95°C | 5s | 40× |
| 60°C* | 5s# |
#Alternatively, set as the minimal time allowed by your Quantitative real-time PCR thermocycler.
Data Analysis
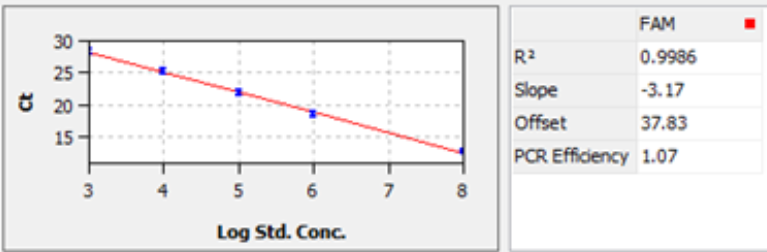
1. Once the qPCR program is finished, the Ct values will generally be calculated in the default threshold (example shown in figure 1).
2. Export the Ct values into Microsoft Excel or other data analysis software. Determine average Ct Values for each triplicate samples. All Ct values should be below that of the NTC.
3. Generate a standard curve to demonstrate linear correlation between Ct values and the standard titer (log scale), like example shown in figure 2. R2 is required to above 0.98, while PCR efficiency 0.90~1.10.
4. Plot the unknown samples Ct values against linear correlation formulation to obtain a equivalent pfu/mL value for each sample.